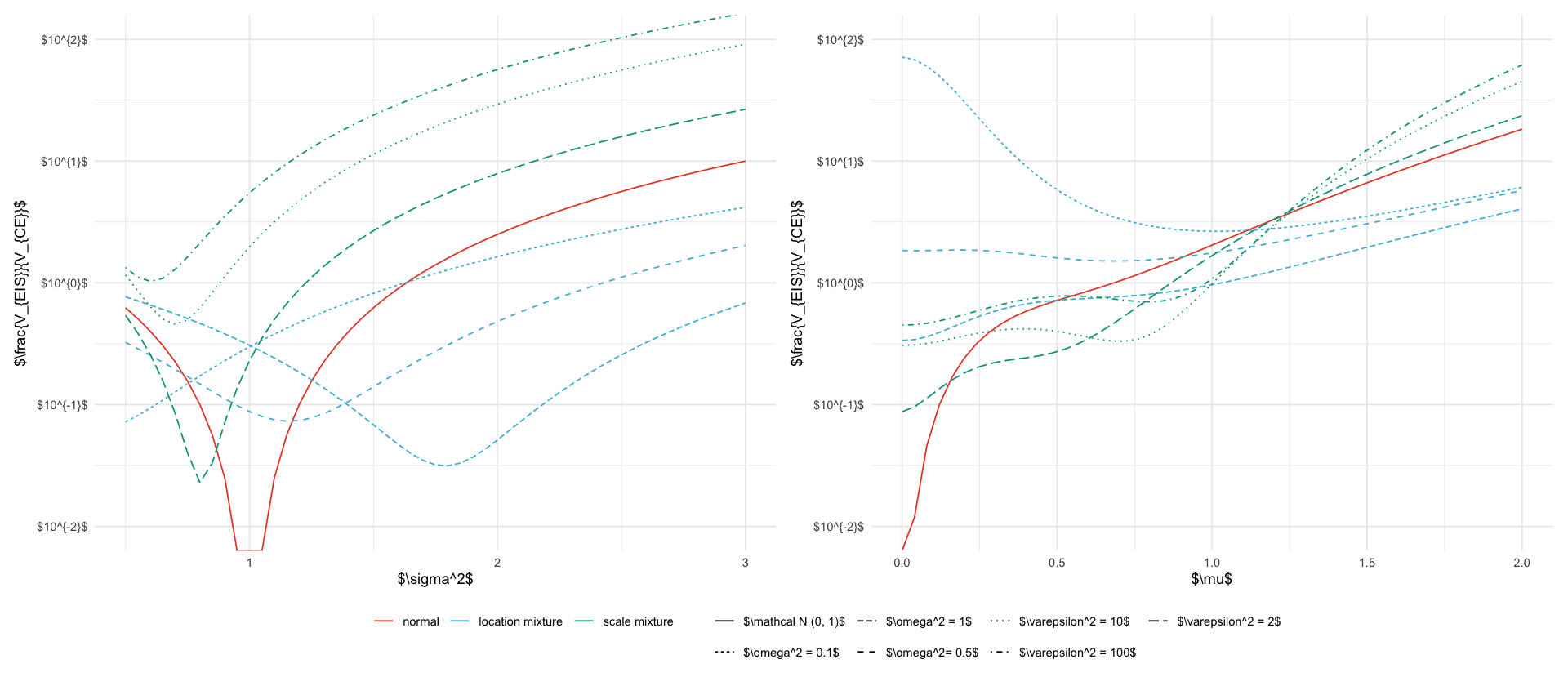
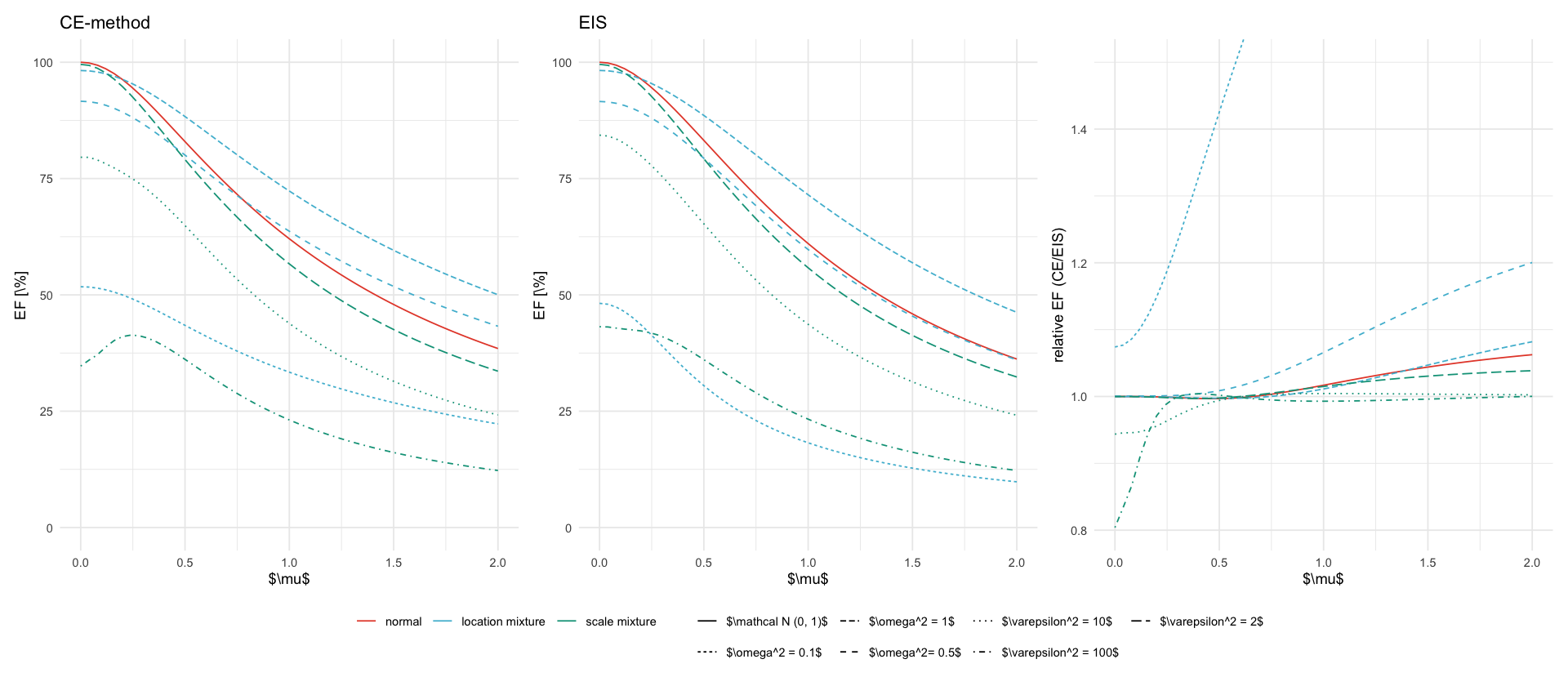
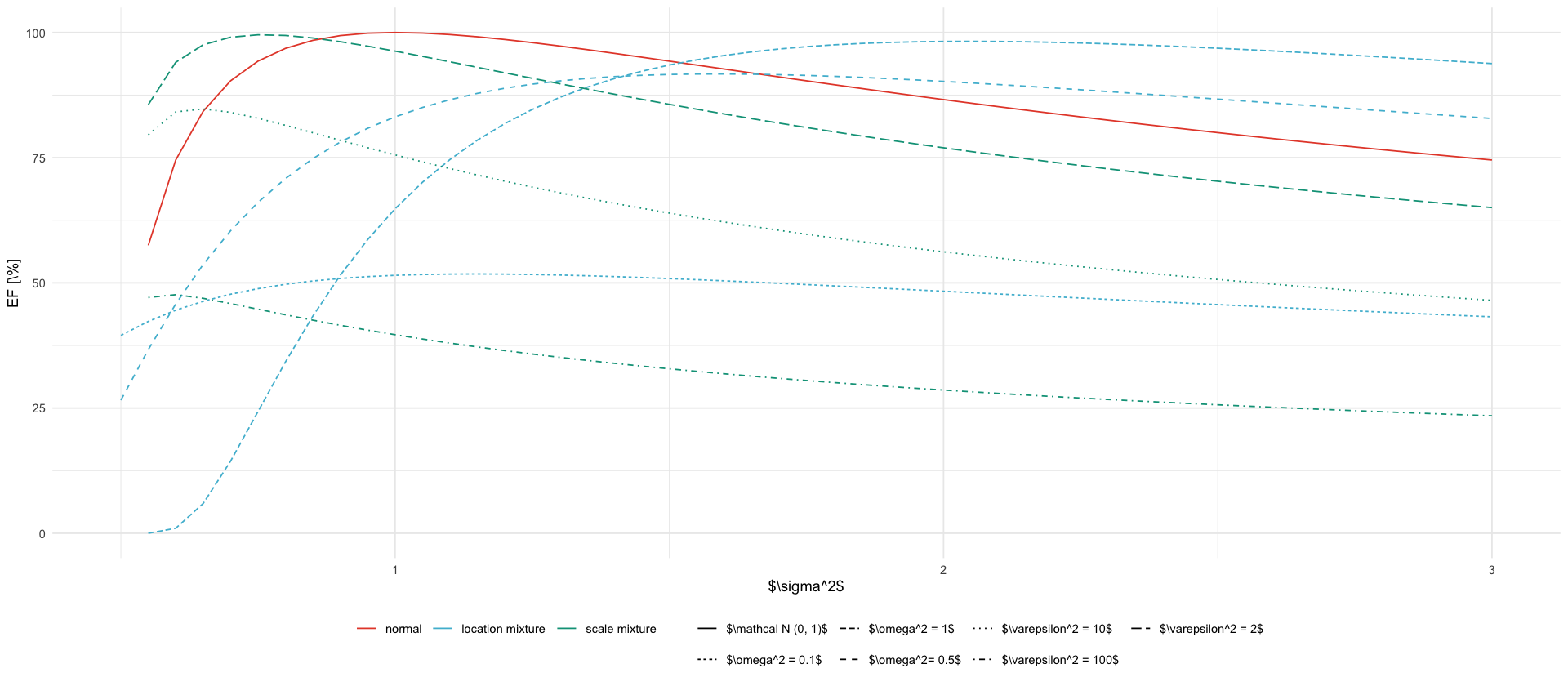
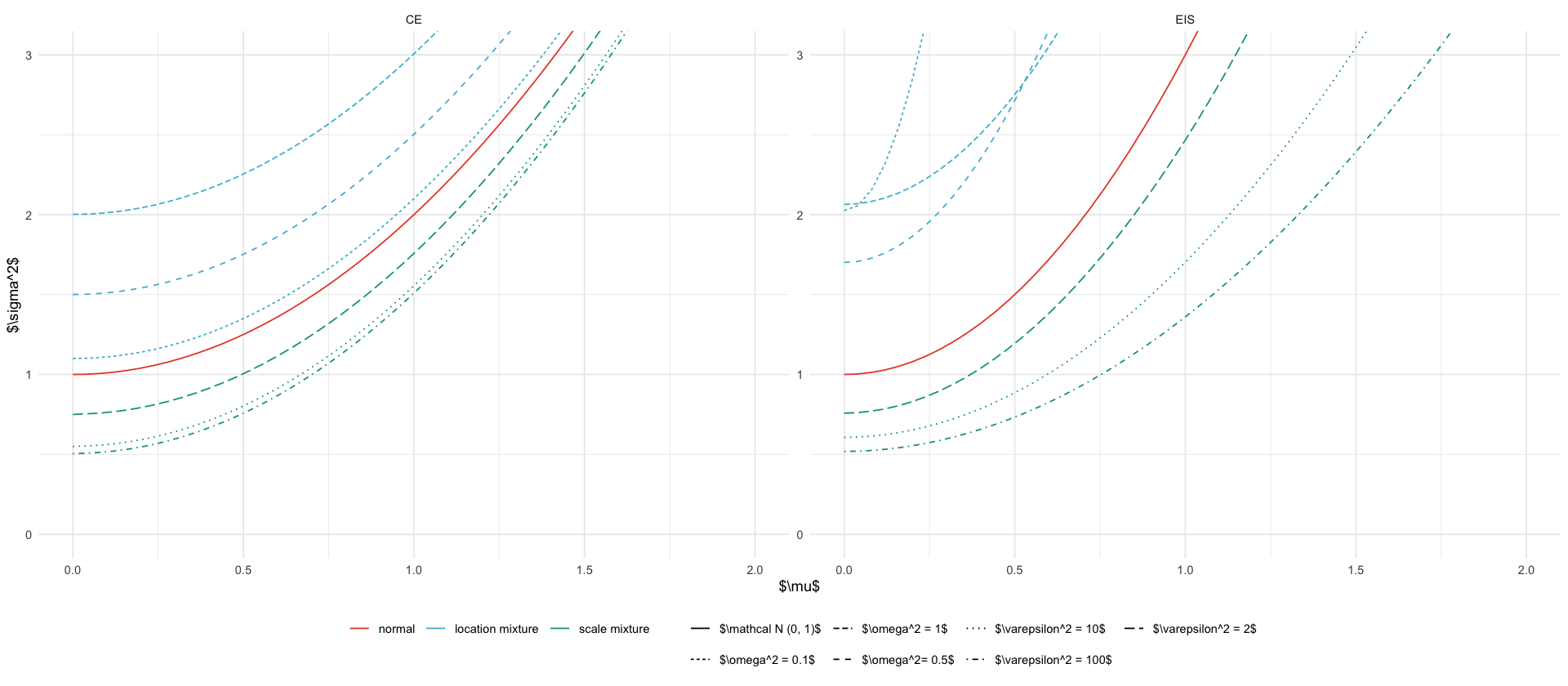
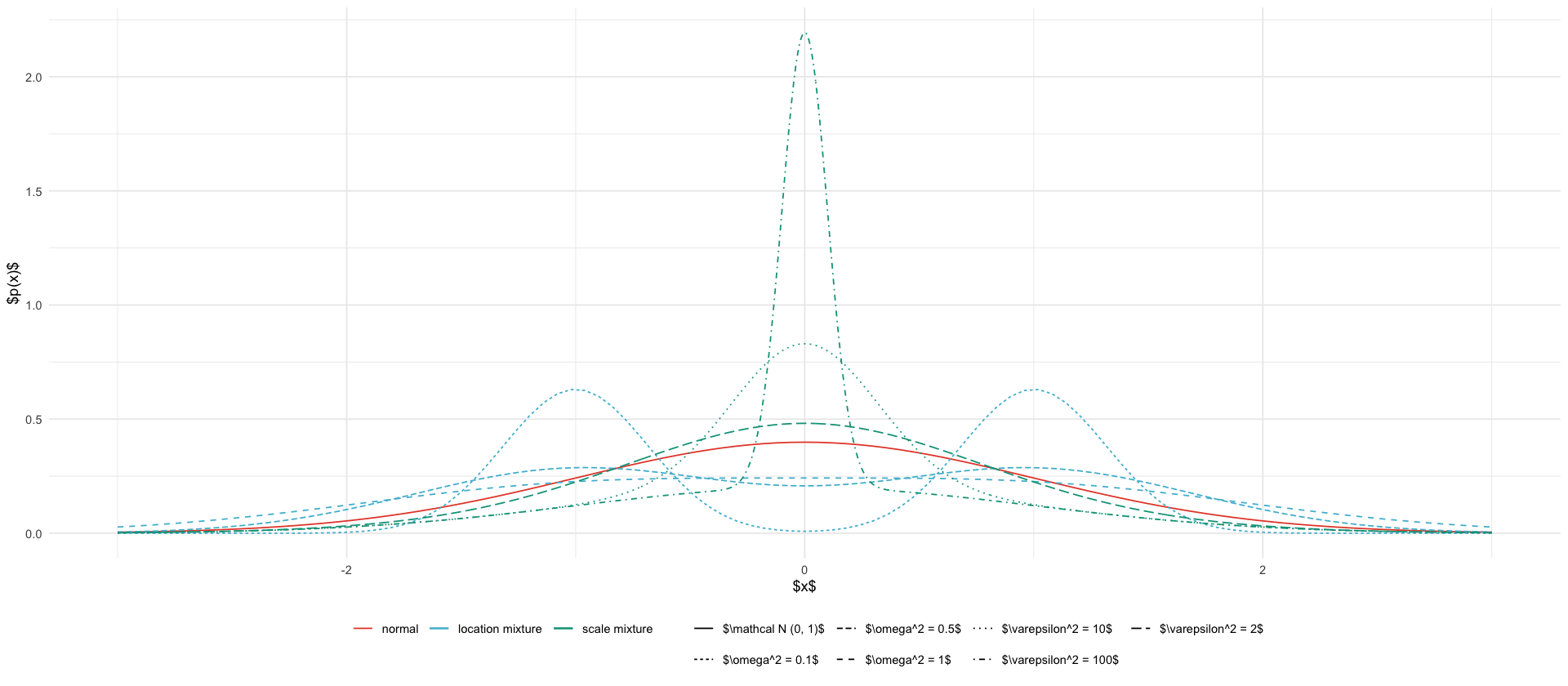
types <- c("normal", "location mixture", "scale mixture")
to_type <- function(label) factor(label, levels = types)
# Define the location mixture function
location_mixture <- function(x, omega2) {
0.5 * dnorm(x, mean = -1, sd = sqrt(omega2)) +
0.5 * dnorm(x, mean = 1, sd = sqrt(omega2))
}
omega2s <- c(0.1, 0.5, 1.0)
loc_geoms <- map(
omega2s,
~geom_function(fun = function(x) location_mixture(x, omega2 = .x), aes(color=to_type("location mixture"), linetype=paste0("$\\omega^2 = ", .x, "$")))
)
# Define the scale mixture function
scale_mixture <- function(x, eps2) {
.5 * dnorm(x, mean=0, sd=1) + .5 * dnorm(x, mean=0, sd=sqrt(1/eps2))
}
eps2s <- c(2, 10, 100)
scale_geoms <- map(
eps2s,
~geom_function(fun = function(x) scale_mixture(x, eps2 = .x), aes(color=to_type("scale mixture"), linetype=paste0("$\\varepsilon^2 = ", .x, "$")), n= 1001)
)
ggplot() +
geom_function(fun = dnorm, aes(color = to_type("normal"), linetype = "$\\mathcal N (0, 1)$")) +
loc_geoms +
scale_geoms +
xlim(-3, 3) +
labs(color = "Distribution", linetype = "Parameter") +
scale_color_discrete(name = "") +
scale_linetype_discrete(name = "") +
theme_minimal() +
theme(legend.position = "bottom") +
labs(x = "$x$", y = "$p(x)$", color = "target", linetype = "parameter")
ggsave_tikz(here("tikz/targets.tex"))