library(here)
source(here("setup.R"))here() starts at /Users/stefan/workspace/work/phd/thesis
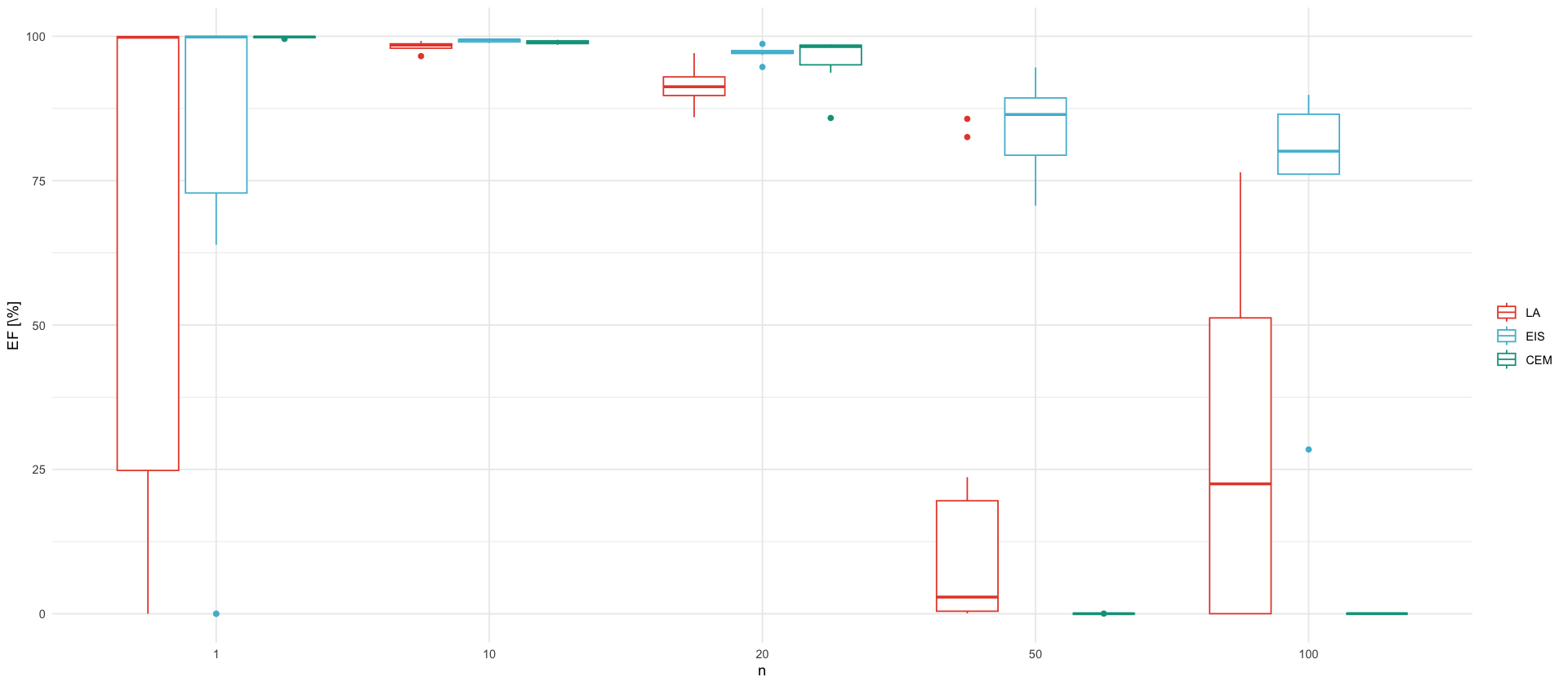
unique_n <- df_ef$n %>% unique() %>% sort()
p_ef <- df_ef %>%
select(n, LA=EF_LA, CEM=EF_CEM, EIS=EF_MEIS) %>%
pivot_longer(-n, names_to = "Method", values_to = "EF") %>%
mutate(Method = factor(Method, levels=c("LA", "EIS", "CEM"))) %>%
ggplot(aes(x = factor(n), y = EF, color = Method, group=interaction(factor(n), Method))) +
geom_boxplot() +
labs(x = "n", y = "EF [\\%]", color="")
ggsave_tikz(
here("tikz/ssm_comparison_efficiency_factor.tex"),
plot = p_ef
)
p_efWarning message: “Removed 26 rows containing non-finite outside the scale range (`stat_boxplot()`).”
Warning message: “Removed 26 rows containing non-finite outside the scale range (`stat_boxplot()`).”
| n | N_samples | N_iter | DET_CEM | DET_MEIS | ARE |
|---|---|---|---|---|---|
| <dbl> | <dbl> | <dbl> | <dbl> | <dbl> | <dbl> |
| 1 | 10000 | 1000 | 1.250198e-21 | 7.362585e-21 | 1.698042e-01 |
| 1 | 10000 | 1000 | 2.131546e-19 | 7.124898e-18 | 2.991686e-02 |
| 1 | 10000 | 1000 | 1.520778e-20 | 6.692526e-20 | 2.272353e-01 |
| 1 | 10000 | 1000 | 6.007386e-17 | 6.660607e-11 | 9.019277e-07 |
| 1 | 10000 | 1000 | 6.624824e-22 | 3.177769e-22 | 2.084741e+00 |
| 1 | 10000 | 1000 | 1.151610e-21 | 3.231204e-21 | 3.564029e-01 |
| 1 | 10000 | 1000 | 3.083249e-23 | 1.207182e-22 | 2.554088e-01 |
| 1 | 10000 | 1000 | 3.179683e-21 | 4.044667e-21 | 7.861420e-01 |
| 1 | 10000 | 1000 | 2.180299e-18 | 8.979118e-15 | 2.428188e-04 |
| 1 | 10000 | 1000 | 1.877286e-20 | 2.278412e-18 | 8.239452e-03 |
| 2 | 10000 | 1000 | 1.927399e-28 | 1.079899e-28 | 1.784796e+00 |
| 2 | 10000 | 1000 | 1.144634e-32 | 2.888226e-32 | 3.963103e-01 |
| 2 | 10000 | 1000 | 6.159994e-29 | 7.289892e-29 | 8.450048e-01 |
| 2 | 10000 | 1000 | 1.429030e-31 | 2.081389e-32 | 6.865755e+00 |
| 2 | 10000 | 1000 | 5.423362e-30 | 7.953996e-30 | 6.818413e-01 |
| 2 | 10000 | 1000 | 5.877860e-32 | 6.436372e-32 | 9.132256e-01 |
| 2 | 10000 | 1000 | 5.379362e-33 | 1.062051e-32 | 5.065069e-01 |
| 2 | 10000 | 1000 | 4.887804e-31 | 5.855831e-30 | 8.346902e-02 |
| 2 | 10000 | 1000 | 2.035986e-32 | 9.830573e-32 | 2.071076e-01 |
| 2 | 10000 | 1000 | 7.752964e-33 | 9.875676e-34 | 7.850566e+00 |
| 5 | 10000 | 1000 | 1.306769e-54 | 3.883234e-60 | 3.365157e+05 |
| 5 | 10000 | 1000 | 7.110526e-50 | 1.654847e-59 | 4.296787e+09 |
| 5 | 10000 | 1000 | 3.682259e-69 | 7.342128e-71 | 5.015248e+01 |
| 5 | 10000 | 1000 | 1.271453e-57 | 3.366841e-62 | 3.776399e+04 |
| 5 | 10000 | 1000 | 1.589256e-62 | 2.441037e-65 | 6.510576e+02 |
| 5 | 10000 | 1000 | 7.400743e-67 | 5.401150e-68 | 1.370216e+01 |
| 5 | 10000 | 1000 | 1.671573e-55 | 2.222286e-61 | 7.521860e+05 |
| 5 | 10000 | 1000 | 6.099313e-58 | 1.303119e-63 | 4.680551e+05 |
| 5 | 10000 | 1000 | 9.018652e-68 | 1.924074e-70 | 4.687269e+02 |
| 5 | 10000 | 1000 | 2.414396e-61 | 8.786312e-65 | 2.747906e+03 |
| 10 | 10000 | 1000 | 1.052184e-139 | -3.827550e-153 | -2.748974e+13 |
| 10 | 10000 | 1000 | -1.867709e-124 | 5.903577e-146 | -3.163691e+21 |
| 10 | 10000 | 1000 | -7.623069e-129 | 1.299750e-146 | -5.865026e+17 |
| 10 | 10000 | 1000 | 3.335174e-127 | -7.751308e-148 | -4.302724e+20 |
| 10 | 10000 | 1000 | -2.491652e-142 | -3.458454e-152 | 7.204527e+09 |
| 10 | 10000 | 1000 | -9.698239e-153 | 2.342988e-159 | -4.139262e+06 |
| 10 | 10000 | 1000 | -4.840100e-124 | 7.639541e-146 | -6.335590e+21 |
| 10 | 10000 | 1000 | -8.222822e-109 | -4.968050e-143 | 1.655141e+34 |
| 10 | 10000 | 1000 | -5.209369e-142 | -7.930262e-154 | 6.568975e+11 |
| 10 | 10000 | 1000 | -7.507561e-142 | 4.376430e-155 | -1.715453e+13 |
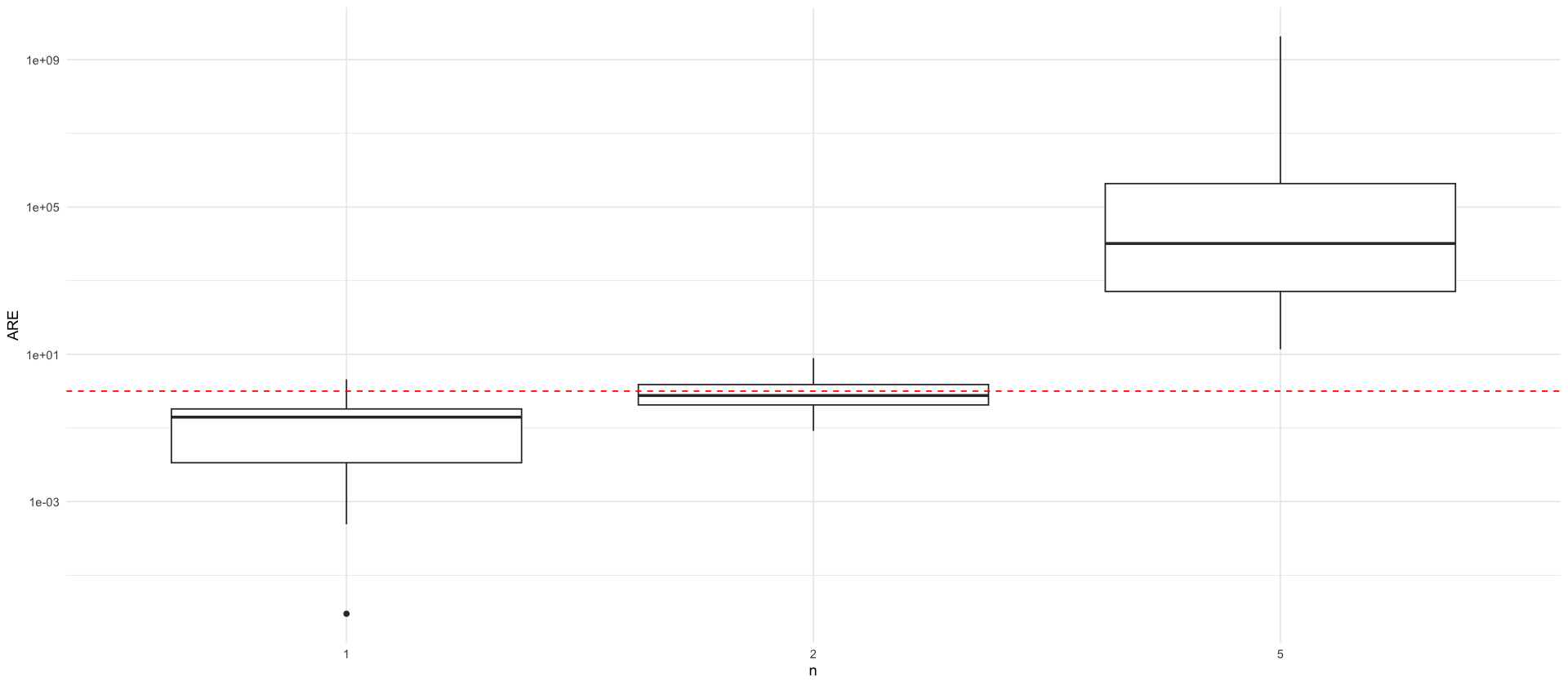
df_are <- read_csv(here("data/figures/are_meis_cem_ssms.csv"))
p_are <- df_are %>%
filter(n <= 5) %>%
ggplot(aes(x = factor(n), y = ARE)) +
geom_boxplot() +
labs(x = "n", y = "ARE", color="") +
geom_hline(yintercept = 1, linetype="dashed", color = "red") +
scale_y_log10()
ggsave_tikz(
here("tikz/ssm_comparison_asymptotic_variance.tex"),
plot = p_are
)
p_are